Microarray based genotyping is an inexpensive and powerful tool to characterize genetic variation. High-density genotyping microarrays are commercially available for humans, economically important livestock and model organisms. However, they have not been available previously for the laboratory mouse, the premier mammalian model organism for biomedical research. Here we describe a custom high-density mouse genotyping array. The Mouse Diversity array was designed to capture the known genetic variation present in the laboratory mouse. It contains 623,124 SNPs distributed across the 19 mouse autosomes, the sex chromosomes, and the mitochondria with a median spacing of one SNP every 1,411 bp in the nuclear genome. The array also contains 916,269 invariant probes that are targeted to functional elements and regions of the genome known to harbor segmental duplications. The nature of the probes opens the door to a variety of novel applications including the characterization of copy number variation, allele specific gene expression and DNA methylation. Performance of the array based on call rate, replication and concordance with previously known genotypes is exceptional. The content-rich Mouse Diversity array provides a critical new tool for mouse genetics including the possibility of extending the successes of genome-wide association studies in humans to the mouse.
Funny comment -- This may be the last chip we do. The economics tell us the line is still below for chips, but sequencing is getting cheaper.
History: people catching mice, trading them, etc. Bottlenecks and all sort of artificial effects.
Diversity 11 classical inbred strains: some chromosomal regions with extremely low diversity. Is this petness? Longevity/Fecundity?
Problem with ascertainment bias: 623124 phylogenetically informative SNPs with known ascertainment
Collaborative Cross -- 8
M. musculus lines -- each contributing equally to the final "line". All inbred by now.
Phenotypes of intermediate CCs: e.g. voluntary exercise goes from 0 miles per day to 18 miles per day.
With 2 different inbred parents, we get complex children but with theoretically predictable phenotype, which means doing GWAS with phenotypes "a la carte".
Also, just by mixing genomes, creating diversity that was not in the parents: very useful novel phenotypic diversity.
Resolution is 7x better. CC will not be GWAS-level, but on the order of 10 genes or MB level resolution. Possibly gene level resolution in 10 generations. Always a mapping resolution panel and a validation panel, going back to the inbred lines.
Selection strictly by random number. Maintaining the diversity is good, lucky because natural selection already took a toll on the original strains.
Done in a way to maximise diversity, not to mimic human population structure. Deep reservoir of diversity for studying phenotypes.
A-Male/B-female crosses and compare to A-Female/B-Male in terms of sex-related epigenetics and other studies.
Some strains will die out along the way, but in 5-10 years should get a lot of info out of it.
Hyuna Yang a lot of array work.
David Aylor pop.struct.
Published mouse distances:
Mus cervicolor - Mus crociduroides = 7.60 MYA
Mus cervicolor - Mus haussa = 6.60 MYA
Mus cervicolor - Mus indutus = 6.60 MYA
Mus cervicolor - Mus mattheyi = 6.60 MYA
Mus cervicolor - Mus minutoides = 6.60 MYA
Mus cervicolor - Mus musculoides = 6.60 MYA
Mus cervicolor - Mus musculus = 4.80 MYA
Mus cervicolor - Mus pahari = 7.60 MYA
Mus cervicolor - Mus platythrix = 7.10 MYA
Mus cervicolor - Mus setulosus = 6.60 MYA
Mus cervicolor - Mus spretus = 4.80 MYA
Mus crociduroides - Mus haussa = 7.60 MYA
Mus crociduroides - Mus indutus = 7.60 MYA
Mus crociduroides - Mus mattheyi = 7.60 MYA
Mus crociduroides - Mus minutoides = 7.60 MYA
Mus crociduroides - Mus musculoides = 7.60 MYA
Mus crociduroides - Mus musculus = 7.60 MYA
Mus crociduroides - Mus pahari = 3.40 MYA
Mus crociduroides - Mus platythrix = 7.60 MYA
Mus crociduroides - Mus setulosus = 7.60 MYA
Mus crociduroides - Mus spretus = 7.60 MYA
Mus haussa - Mus indutus = 3.20 MYA
Mus haussa - Mus mattheyi = 2.60 MYA
Mus haussa - Mus minutoides = 3.20 MYA
Mus haussa - Mus musculoides = 3.20 MYA
Mus haussa - Mus musculus = 6.60 MYA
Mus haussa - Mus pahari = 7.60 MYA
Mus haussa - Mus platythrix = 7.10 MYA
Mus haussa - Mus setulosus = 4.00 MYA
Mus haussa - Mus spretus = 6.60 MYA
Mus indutus - Mus mattheyi = 3.20 MYA
Mus indutus - Mus minutoides = 2.50 MYA
Mus indutus - Mus musculoides = 2.50 MYA
Mus indutus - Mus musculus = 6.60 MYA
Mus indutus - Mus pahari = 7.60 MYA
Mus indutus - Mus platythrix = 7.10 MYA
Mus indutus - Mus setulosus = 4.00 MYA
Mus indutus - Mus spretus = 6.60 MYA
Mus mattheyi - Mus minutoides = 3.20 MYA
Mus mattheyi - Mus musculoides = 3.20 MYA
Mus mattheyi - Mus musculus = 6.60 MYA
Mus mattheyi - Mus pahari = 7.60 MYA
Mus mattheyi - Mus platythrix = 7.10 MYA
Mus mattheyi - Mus setulosus = 4.00 MYA
Mus mattheyi - Mus spretus = 6.60 MYA
Mus minutoides - Mus musculoides = 1.60 MYA
Mus minutoides - Mus musculus = 6.60 MYA
Mus minutoides - Mus pahari = 7.60 MYA
Mus minutoides - Mus platythrix = 7.10 MYA
Mus minutoides - Mus setulosus = 4.00 MYA
Mus minutoides - Mus spretus = 6.60 MYA
Mus musculoides - Mus musculus = 6.60 MYA
Mus musculoides - Mus pahari = 7.60 MYA
Mus musculoides - Mus platythrix = 7.10 MYA
Mus musculoides - Mus setulosus = 4.00 MYA
Mus musculoides - Mus spretus = 6.60 MYA
Mus musculus - Mus pahari = 7.60 MYA
Mus musculus - Mus platythrix = 7.10 MYA
Mus musculus - Mus setulosus = 6.60 MYA
Mus musculus - Mus spretus = 2.30 MYA
Mus pahari - Mus platythrix = 7.60 MYA
Mus pahari - Mus setulosus = 7.60 MYA
Mus pahari - Mus spretus = 7.60 MYA
Mus platythrix - Mus setulosus = 7.10 MYA
Mus platythrix - Mus spretus = 7.10 MYA
Mus setulosus - Mus spretus = 6.60 MYA
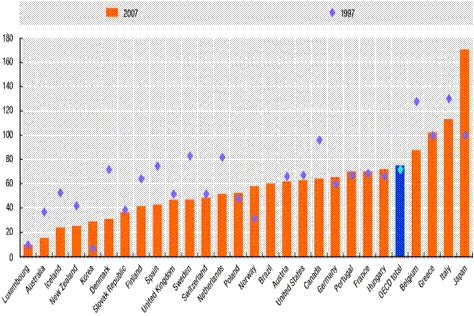 OECD Government debt as % of GDP
OECD Government debt as % of GDP














































